GuideMaker
GuideMaker: Software to design CRISPR-Cas guide RNA pools in non-model genomes 🦠 🧬
CRISPR-Cas systems have expanded the possibilities for gene editing in bacteria and eukaryotes. There are many excellent tools for designing the CRISPR-Cas guide RNAs for model organisms with standard Cas enzymes. GuideMaker is intended as a fast and easy-to-use design tool for atypical projects with 1) non-standard Cas enzymes, 2) non-model organisms, or 3) projects that need to design a panel of guide RNAs (gRNA) for genome-wide screens.
GuideMaker can rapidly design gRNAs for gene targets across the genome from a degenerate protospacer adjacent motif (PAM) and a GenBank file or Fasta and GFF/GTF file. The tool applies Hierarchical Navigable Small World (HNSW) graphs to speed up the comparison of guide RNAs enabling the user to design gRNAs for all genes for a typical bacterial genome and PAM sequence in about 1-2 minutes on a laptop.
GuideMaker enables the rapid design of genome-wide CRISPR/Cas gene function studies in non-model organisms with any Cas enzyme. While GuideMaker is designed with prokaryotic genomes in mind, it can process smaller eukaryotic genomes as well. GuideMaker is available as command-line software and as a web application at https://guidemaker.app.scinet.usda.gov and in the CyCverse Discovery Environment.
Methods to access GuideMaker
GuideMaker can be easily accessed via:
- Web Application
- CyVerse Discovery Environment
- Command Line
- Local Web Application
NOTE: *Our web application runs on a small server instance and is primarily designed for the lower-memory requirements bacterial genomes. We recommend that users run larger genomes on the CyCverse Discovery Environment or run GuideMaker locally as a command-line or web browser-based application.
| 1.Web Application | 2.CyCverse Discovery Environment |
|---|---|
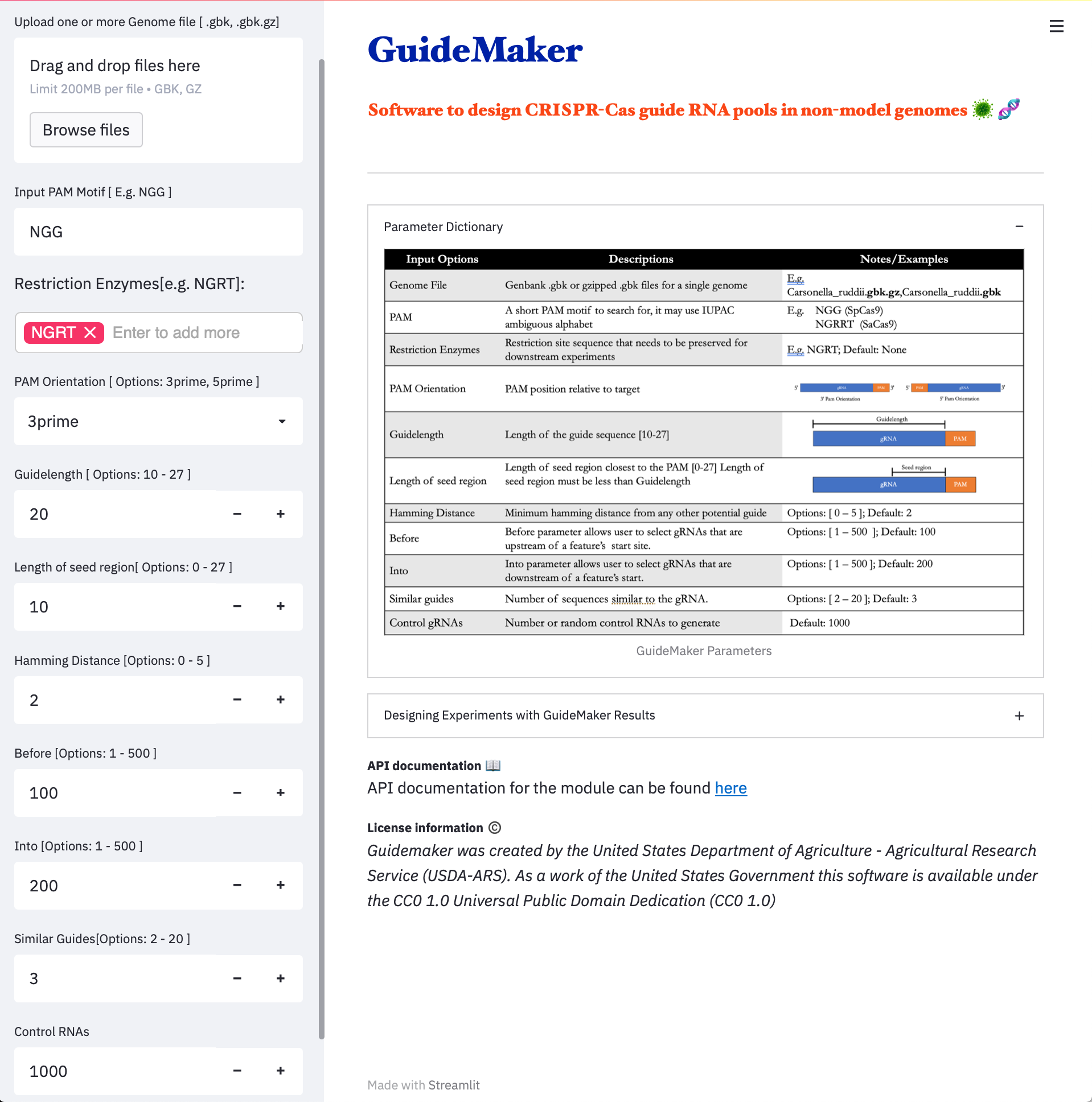 | 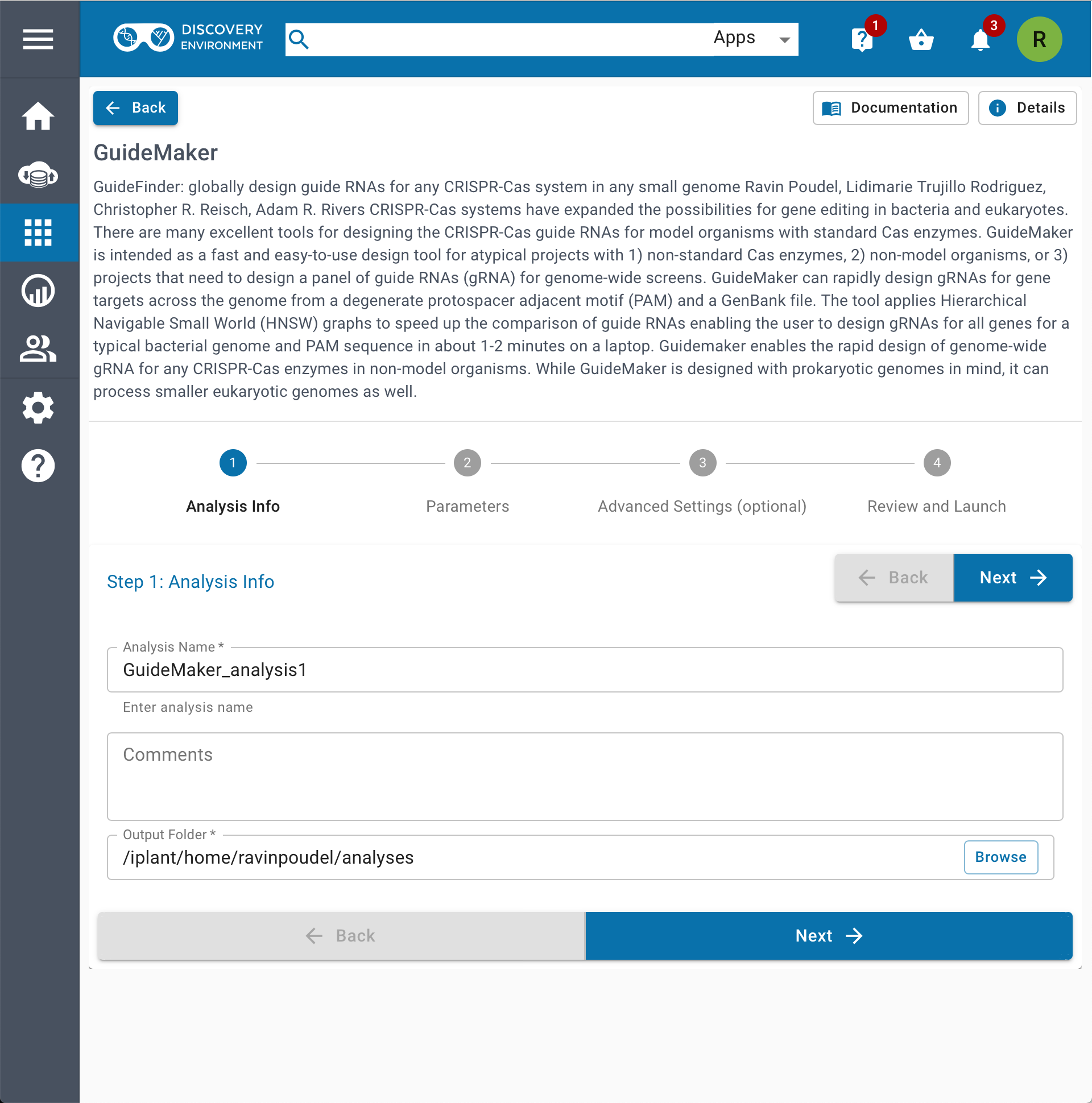 |
3.Command Line
GuideMaker can be installed from:
Docker image (prefered method)
Guidemaker is available at the Github Container Registry (https://github.com/orgs/USDA-ARS-GBRU/packages?repo_name=GuideMaker)
# AVX Version
docker pull ghcr.io/usda-ars-gbru/guidemaker-avx
#Non-AVX version
docker pull ghcr.io/usda-ars-gbru/guidemaker-noavx
Bioconda
# Create a conda environment and install GuideMaker via Bioconda.
mamba create --strict-channel-priority --override-channels --channel conda-forge --channel bioconda --channel defaults --name gmenv guidemaker
# Activate conda env
mamba activate gmenv
# Test the installation
guidemaker -h
Github
# Create a conda environment and install and pybedtools
mamba create -n gmenv python=3.9 pybedtools=0.9.1
conda activate gmenv
git clone https://github.com/USDA-ARS-GBRU/GuideMaker.git
cd GuideMaker
pip install .
# check if the installation works
guidemaker -h
Dependencies
pybedtoolsNMSLibBiopythonPandasStreamlit for webappaltair for plotting
Command Line Usage
usage: guidemaker [-h] [--genbank GENBANK [GENBANK ...]] [--fasta FASTA [FASTA ...]] [--gff GFF [GFF ...]] --pamseq PAMSEQ --outdir OUTDIR [--raw_output_only]
[--pam_orientation {5prime,3prime}] [--guidelength [10-27]] [--lsr [0-27]] [--dtype {hamming,leven}] [--dist [0-5]] [--before [1-500]] [--into [1-500]]
[--knum [2-20]] [--controls [0-100000]] [--threads THREADS] [--log LOG] [--tempdir TEMPDIR] [--restriction_enzyme_list [RESTRICTION_ENZYME_LIST ...]]
[--attribute_key ATTRIBUTE_KEY] [--filter_by_attribute [FILTER_BY_ATTRIBUTE ...]] [--doench_efficiency_score] [--cfd_score] [--keeptemp] [--plot] [--config CONFIG]
[-V]
GuideMaker: Software to design gRNAs pools in non-model genomes and CRISPR-Cas systems
optional arguments:
-h, --help show this help message and exit
--genbank GENBANK [GENBANK ...], -i GENBANK [GENBANK ...]
One or more genbank .gbk or gzipped .gbk files for a single genome. Provide this or GFF/GTF and fasta files
--fasta FASTA [FASTA ...], -f FASTA [FASTA ...]
One or more fasta or gzipped fasta files for a single genome. If using a fasta, a GFF/GTF file must also be provided but not a genbank file.
--gff GFF [GFF ...], -g GFF [GFF ...]
One or more GFF or GTF files (optionally gzipped) for a single genome. If using a GFF/GTF a fasta file must also be provided but not a genbank file.
--pamseq PAMSEQ, -p PAMSEQ
A short PAM motif to search for, it may use IUPAC ambiguous alphabet
--outdir OUTDIR, -o OUTDIR
The directory for data output
--raw_output_only if selected only the raw guide RNAs their positions will be returned the meet lsr and dist criteria
--pam_orientation {5prime,3prime}, -r {5prime,3prime}
The PAM position relative to the target: 5prime: [PAM][target], 3prime: [target][PAM]. For example, SpCas9 is 3prime. Default: '3prime'.
--guidelength [10-27], -l [10-27]
Length of the guide sequence. Default: 20.
--lsr [0-27] Length of a seed region near the PAM site required to be unique. Default: 10.
--dtype {hamming,leven}
Select the distance type. Default: hamming.
--dist [0-5] Minimum edit distance from any other potential guide. Default: 2.
--before [1-500] keep guides this far in front of a feature. Default: 100.
--into [1-500] keep guides this far inside (past the start site)of a feature. Default: 200.
--knum [2-20] how many sequences similar to the guide to report. Default: 5.
--controls [0-100000]
Number of random control RNAs to generate. Default: 1000.
--threads THREADS The number of cpu threads to use. Default: 2
--log LOG Log file
--tempdir TEMPDIR The temp file directory
--restriction_enzyme_list [RESTRICTION_ENZYME_LIST ...]
List of sequence representing restriction enzymes. Default: None.
--attribute_key ATTRIBUTE_KEY
the attribute key in column 9 of the GFF/GTF file to use for filtering. Default: ID
--filter_by_attribute [FILTER_BY_ATTRIBUTE ...]
List of locus ids. Default: None.
--doench_efficiency_score
On-target scoring from Doench et al. 2016 - only for NGG PAM and guidelength=25: Default: None.
--cfd_score CFD score for assessing off-target activity of gRNAs with NGG pam: Default: None.
--keeptemp Option to keep intermediate files be kept
--plot Option to create GuideMaker plots
--config CONFIG Path to YAML formatted configuration file, default is /Users/rivers/Documents/guidemaker/guidemaker/data/config_default.yaml
-V, --version show program's version number and exit
To run the web app locally, in your terminal run:
-----------------------------------------------------------------------
streamlit run /your/path/to/guidemaker/guidemaker/data/app.py
-----------------------------------------------------------------------
Examples
Use case: Make 20 nucleotide guide sequences for SpCas9 (NGG) in the bacterium Carsonela ruddii. The length of the seed region near the PAM required to be unique in each guide is 11 nucleotides.
guidemaker \
-i tests/test_data/Carsonella_ruddii.gbk \
-p NGG \
--pam_orientation 3prime \
--guidelength 20 \
--lsr 11 \
-o OUTDIR \
--doench_efficiency_score \
--threads 2
4. Running Web App locally
To run the web app locally, you first need to complete the command line installation described above.
If the path of the app.py differs from the one displayed below, you can locate the path by first running guidemaker --help. Script to run the web app locally is available at the bottom of the help command output. If you need to upload larger genomes you can change the server.maxUploadSize parameter which id in MB.
streamlit run /[user path prefix]/guidemaker/data/app.py --server.maxUploadSize 500

Running the docker image
Command line usage: on a computer with AVX (most modern computers) you can run these commands:
docker pull docker pull ghcr.io/usda-ars-gbru/guidemaker-avx
docker run -it ghcr.io/usda-ars-gbru/guidemaker-avx guidemaker -h
Web App usage: on a computer with AVX (most modern computers) you can run these commands:
docker pull docker pull ghcr.io/usda-ars-gbru/guidemaker-avx
docker run -p8501:8501 -it ghcr.io/usda-ars-gbru/guidemaker-avx
Now if you open your browser you can access the app at http://127.0.0.1:8501
Using GuideMaker's results
This section provides information on how to use GuideMaker's results to create a molecular protocol for pooled CRISPR screens.
Pooled CRISPR Experiments
Experiments that target the entire genome, or many genes at once, are typically performed in pooled experiments where 100-100,000+ targets are tested simultaneously. The pooled oligonucleotides for each gRNA are cloned in one batch and used simultaneously in the designed experiment. Each gRNA sequence acts as a barcode that can be quantified with high-throughput sequencing to elucidate each target's relative importance under the experimental conditions.
Vectors for gRNA Cloning
Genome-scale CRISPR experiments require a gRNA vector amenable to high-throughput cloning, most often through Golden Gate cloning, a restriction enzyme-dependent reaction. Plasmids to express gRNA are available from Addgene can be found at the link below, though not all of these are compatible with high-throughput cloning.
Addgene: CRISPR Plasmids - Empty gRNA Vectors
After running GuideMaker, the designed gRNA output can be downloaded and with minor adjustments, the targets can be ordered as oligos for cloning. Pooled oligonucleotides can be purchased from several vendors, including those listed below. Pool sizes vary from 100 to over 200,000 oligonucleotides. Vendor specifications for the number of oligos, oligo length, and cost per bp vary widely. For bacterial genome-scale experiments, as of 2021, Genscript offers pool sizes of 12,472 and 91,766 with up to 79 bp per oligo for list prices of $1600 and $4,000, respectively.
Some example vendors are:
Most pools require amplification before cloning to convert the ssDNA to dsDNA and increase the concentration for efficient cloning. Accordingly, adding a constant region at the 3' end for primer binding is recommended. Sub-pools can also be amplified by adding unique constant regions to some oligos, enabling the large-scale synthesis to be split amongst organisms or specific targets in a single organism. Because Golden Gate cloning utilizes restriction enzymes, filtering gRNA designs with the cognate restriction enzyme recognition sites is necessary, a feature found in GuideMaker. A general protocol for cloning pooled gRNA from synthesized oligonucleotides from IDT is linked below, though similar workflows can be used for pools from other vendors.
- Cloning high-quality CRISPR libraries with oPool Oligo Pools (SYB-10182-PR12/2019)
- Addgene: Guide to Using Pooled Libraries
Pooled CRISPR Data Analysis
After the experiment, the cells are collected and DNA is isolated. The target sequence is then amplified and adaptors for high-throughput sequencing added. Several data analysis pipelines have been developed to identify target sequences over-represented or under-represented in the pool. The manuscript by Wang et al. (2019) provides a protocol for using a high-quality tool with these capabilities.
Wang, B., Wang, M., Zhang, W. et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat Protoc 14, 756–780 (2019). https://doi.org/10.1038/s41596-018-0113-7
Reporting Errors and Suggestions
Open the GuideMaker github repo, navigate to the Issues page and submit an issue to report difficulties, errors, or suggestions for improvements.
GuideMaker Citation
Ravin Poudel, Lidimarie Trujillo Rodriguez, Christopher R Reisch, Adam R Rivers, GuideMaker: Software to design CRISPR-Cas guide RNA pools in non-model genomes, GigaScience, Volume 11, 2022, giac007, DOI: 10.1093/gigascience/giac007
API documentation
API documentation for the module can be found here
License information
GuideMaker was created by the United States Department of Agriculture - Agricultural Research Service (USDA-ARS). As a work of the United States Government, this software is available under the CC0 1.0 Universal Public Domain Dedication (CC0 1.0)
About us
GuideMaker was developed by the USDA Agricultural Research Service, Genomics and Bioinformatics Research Unit group in Gainesville, FL led by Adam Rivers. Check out our other work at https://tinyecology.com.

